Evolution 2021
Evolving towards a specialized state is rare: A case study of passerine nest type
A long standing hypothesis in evolutionary biology is that the evolution of resource specialization is linked to diversification. However, in recent years, advances in comparative methods investigating trait-based differences associated with diversification have allowed more robust tests of this idea and have found mixed support. Thus, it remains unclear whether resource specialization has significant impacts on patterns of diversification. Using state-dependent diversification models, especially with large data sets, has the potential to bring greater clarity to this question. In this work, we test the evolutionary dead end hypothesis by estimating net diversification rate differences associated with nest site specialization among 3,224 species of passerine birds, and their ancestors. In particular, we test whether the adoption of hole-nesting, a nest site specialization that decreases predation, results in reduced diversification rates relative to nesting outside of holes. Further, we examine whether evolutionary transitions to the specialist hole-nesting state have been more frequent than transitions out of hole-nesting. Using hidden-state diversification models that account for background rate heterogeneity, in addition to accounting for the possibility that passerines have had high extinction rates, we found that nest site was not associated with diversification rate differences. Furthermore, contrary to the assumption that specialized states are difficult to reverse, we instead found that transitions out of hole nesting have been relatively frequent, whereas adoptions of hole nesting have been less frequent. These results suggest that competition may limit transitions into hole nesting, but that such competition for a limited resource does not result in limited diversification. In conjunction with other recent studies using robust comparative methods, our results add to growing evidence that evolutionary dead ends are not a typical outcome of resource specialization.
A little more about extinction…
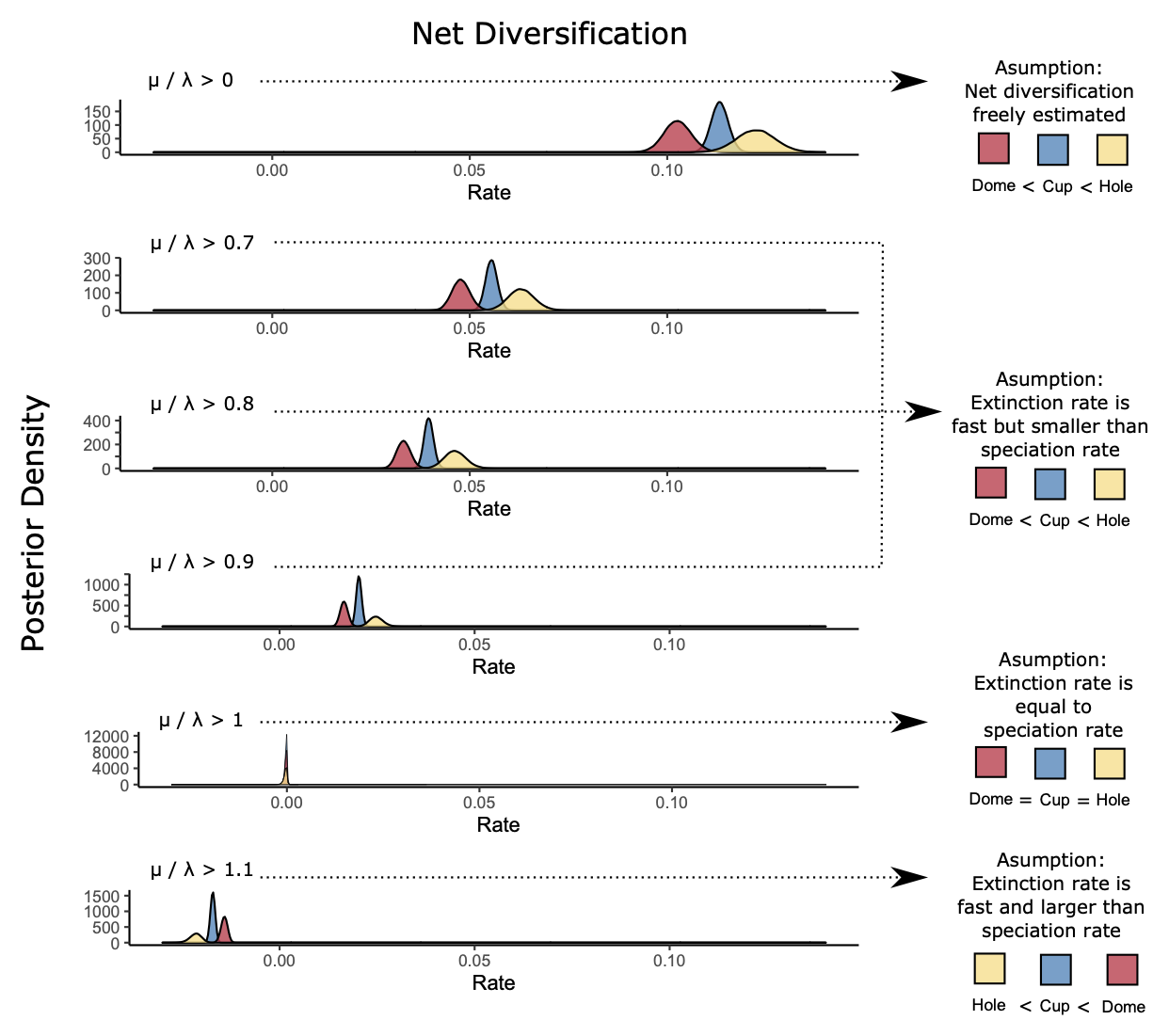
When we assume that the prior distribution of the extinction fraction \(\mu/\lambda\) is enhanced, we obtained the same overall relative inference about differences by state. This is equivalent to assume much more extinction than what traditionally is estimated by diversification models (see Louca and Pennell 2021 preprint about exinction rate estimates being always closer to zero). Even if we don’t recover point estimates in SSE models, we can recover the same critical inference by state. In our case, the state hole (yellow) is the most critical state for diversification (see figure below).
Recent Preprints and Publications
-
Helmstetter, A.J, Glemin, S., Käfer, J., Zenil-Ferguson, R., Sauquet, H., de Boer, H., Dagallier, L.M.J., Mazet, N., Reboud, E.L., Couvreur, T.L.P, Condamine, F.L. Pulled Diversification Rates, Lineages-Through-Time Plots and Modern Macroevolutionary Modelling. bioRxiv 2021.01.04.424672
-
Fumia, N., Rubinoff, D., Zenil-Ferguson, R., Khoury, C.K., Pironon, S., Gore, M.A., Kantar, M.B. Interactions between specific breeding system and ploidy play a critical role in increasing niche adaptability in a global food crop. bioRxiv 2020.09.09.290429
-
Zenil-Ferguson, R., Burleigh, G., Igic, B., Freyman, W.A., Mayrose, I. and Goldberg, E.E. 2019. Interaction between Ploidy, Breeding System, and Lineage Diversification. New Phytologist. pdf supp1 supp 2
-
Ferguson, J.M., Taper, M.L., Zenil-Ferguson, R., Jasieniuk, M. and Maxwell, B., 2019. Incorporating parameter estimability into model selection. Frontiers in Ecology and Evolution, 7, p.427.
-
Rivero, R., Sessa, E.B. and Zenil‐Ferguson, R. 2019. EyeChrom and CCDBcurator: Visualizing chromosome count data from plants. Applications in Plant Sciences. Undergrad research: Visit Eyechrom website
-
Uyeda, J.C., Zenil-Ferguson, R. and Pennell, M.W., 2018.Rethinking phylogenetic comparative methods. Systematic Biology, 67(6), pp.1091-1109.
-
Zenil‐Ferguson, R., Burleigh, J.G. and Ponciano, J.M., 2018. chromploid: An R package for chromosome number evolution across the plant tree of life. Applications in plant sciences, 6(3), p.e1037.
-
Landis, J.B., Bell, C.D., Hernandez, M., Zenil-Ferguson, R., McCarthy, E.W., Soltis, D.E. and Soltis, P.S., 2018. Evolution of floral traits and impact of reproductive mode on diversification in the phlox family (Polemoniaceae). Molecular phylogenetics and evolution, 127, pp.878-890.